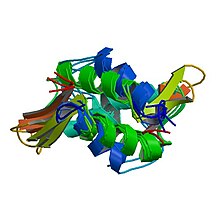
Histone H1
| |||||||||||||||||||||||||||||
Read other articles:

Artikel ini tidak memiliki referensi atau sumber tepercaya sehingga isinya tidak bisa dipastikan. Tolong bantu perbaiki artikel ini dengan menambahkan referensi yang layak. Tulisan tanpa sumber dapat dipertanyakan dan dihapus sewaktu-waktu.Cari sumber: Dinden, Kwadungan, Ngawi – berita · surat kabar · buku · cendekiawan · JSTOR DindenDesaNegara IndonesiaProvinsiJawa TimurKabupatenNgawiKecamatanKwadunganKode pos63283Kode Kemendagri35.21.06.2002 Luas..…

Papuan language of East Timor FatalukuRegionEastern East TimorNative speakers48,000 (2020)[1]regional usageLanguage familyTrans–New Guinea ? West Bomberai ?Timor–Alor–PantarEastern TimorOirata–FatalukuFataluku–RusenuFatalukuOfficial statusRecognised minoritylanguage inEast TimorLanguage codesISO 639-3ddgGlottologfata1247Distribution of Fataluku in East Timor Fataluku (also known as Dagaga, Dagoda', Dagada) is a Papuan language spoken by approximately 37,000 …

Messier 53Messier 53 oleh Teleskop Luar Angkasa Hubble; 3.5′ view Kredit: NASA/STScI/WikiSkyData pengamatan (J2000 epos)KelasV[1]Rasi bintangComa BerenicesAsensio rekta 13j 12m 55.25d[2]Deklinasi +18° 10′ 05.4″[2]Jarak58 kly (18.000 pc)[3]Magnitudo tampak (V)+8.33[4]Dimensi tampak (V)13.0′[5]Karakteristik fisikMassa8,26×105[6] M☉MetalisitasTemplat:Fe/H dexPerkiraan umur12.67 …

Historic church in Missouri, United States United States historic placeSt. Francis Xavier College ChurchU.S. Historic districtContributing propertySt. Louis Landmark The College Church in 2018Show map of St. LouisShow map of MissouriShow map of the United StatesLocationLindell and N. Grand Blvds., St. Louis, MissouriCoordinates38°38′13″N 90°13′59″W / 38.63694°N 90.23306°W / 38.63694; -90.23306Arealess than one acreBuilt1884-1898ArchitectThomas WalshHenry Switz…

Motion picture shot with little to no funding from a major film studio or private investor Low Budget redirects here. For the album, see Low Budget (album). For the song, see Low Budget (song). Part of a series onB movies Low-budget films Hollywood Golden Age 1950s transition Exploitation boom Since the 1980s Z movie vte A low-budget film or low-budget movie is a motion picture shot with little to no funding from a major film studio or private investor. Many independent films are made on low bud…

Walter EuckenIstituto Walter EuckenLahir(1891-01-17)17 Januari 1891Meninggal20 Maret 1950(1950-03-20) (umur 59)KebangsaanGerman EmpireBidangMacroeconomicsMazhabOrdoliberalismKontribusiSocial market economy Walter Eucken (17 Januari 1891 – 20 Maret 1950) adalah seorang ahli ekonomi asal Jerman yang juga dikenal sebagai Bapak Ordoliberalisme. Ia dilahirkan di Jena, Thuringen. Ayahnya adalah salah seorang penerima Penghargaan Nobel pada tahun 1908 dalam bidang Literatur yaitu R…

Amedeo Maiuri Amedeo Maiuri (Veroli, 7 gennaio 1886 – Napoli, 7 aprile 1963) è stato un archeologo italiano. Indice 1 Gli studi e la carriera 2 Gli scavi 3 Alcune pubblicazioni 4 Note 5 Voci correlate 6 Altri progetti 7 Collegamenti esterni Gli studi e la carriera Amedeo Maiuri nacque a Veroli (all'epoca Provincia di Roma, oggi Provincia di Frosinone) da Giuseppe, che in quella città era procuratore, ed Elena Parsi; frequentò il ginnasio presso il collegio scolopio Conti-Gentili di Alatri, …

The neutrality of this article is disputed. Relevant discussion may be found on the talk page. Please do not remove this message until conditions to do so are met. (September 2023) (Learn how and when to remove this template message) 'Adam's Creation Sistine Chapel ceiling' by Michelangelo Religion (when discussed as a virtue) is a distinct moral virtue whose purpose is to render God the worship due to Him as the source of all being and the giver of all good things. As such, in Christianity it i…

Artikel ini sudah memiliki daftar referensi, bacaan terkait, atau pranala luar, tetapi sumbernya belum jelas karena belum menyertakan kutipan pada kalimat. Mohon tingkatkan kualitas artikel ini dengan memasukkan rujukan yang lebih mendetail bila perlu. (Pelajari cara dan kapan saatnya untuk menghapus pesan templat ini) Chuck NorrisNorris pada tahun 2015LahirCarlos Ray Norris10 Maret 1940 (umur 84) Ryan, Oklahoma, Amerika SerikatSuami/istriGena O'Kelley (1998-sekarang)Diane Holechek (1958-19…

Jamaican musician (1942–2021) Hugh Roy redirects here. For the cricketer, see Hugh Roy (cricketer). U-RoyODBackground informationBirth nameEwart BeckfordAlso known asThe OriginatorHugh RoyThe TeacherBorn(1942-09-21)21 September 1942Jones Town, JamaicaDied17 February 2021(2021-02-17) (aged 78)Kingston, JamaicaGenresReggaerocksteadydancehalldubOccupation(s)Singer, songwriterYears active1961–2021LabelsTreasure Isle, Duke Reid, VirginMusical artist Ewart Beckford OD (21 September 1942 – 1…

Untuk orang lain dengan nama sama, lihat James Black. Biografi ini tidak memiliki sumber tepercaya sehingga isinya tidak dapat dipastikan. Bantu memperbaiki artikel ini dengan menambahkan sumber tepercaya. Materi kontroversial atau trivial yang sumbernya tidak memadai atau tidak bisa dipercaya harus segera dihapus.Cari sumber: James Whyte Black – berita · surat kabar · buku · cendekiawan · JSTOR (Pelajari cara dan kapan saatnya untuk menghapus pesan templ…

Сибирский горный козёл Научная классификация Домен:ЭукариотыЦарство:ЖивотныеПодцарство:ЭуметазоиБез ранга:Двусторонне-симметричныеБез ранга:ВторичноротыеТип:ХордовыеПодтип:ПозвоночныеИнфратип:ЧелюстноротыеНадкласс:ЧетвероногиеКлада:АмниотыКлада:СинапсидыКласс:…

Italian painter Marcello Venusti, Holy Family, oil on wood, 43.2 x 28.6 cm, c. 1565. National Gallery, London Marcello Venusti (1512 – 15 October 1579)[1] was an Italian Mannerist painter active in Rome in the mid-16th century. Native to Mazzo di Valtellina near Como, he was reputed to have been a pupil of Perino del Vaga. He is known for a scaled copy in oils (now Museo di Capodimonte, Naples) of Michelangelo's Last Judgement in the Sistine Chapel, commissioned by Cardinal Alessandro …

1964 film by B. R. Panthulu Muradan MuthuTheatrical release posterDirected byB. R. PanthuluScreenplay byM. S. SolaimalaiProduced byB. R. PanthuluStarringSivaji GanesanDevikaCinematographyV. RamamurthiEdited byR. DevarajanMusic byT. G. LingappaProductioncompanyPadmini PicturesRelease date 3 November 1964 (1964-11-03) CountryIndiaLanguageTamil Muradan Muthu (transl. Rogue Muthu) is a 1964 Indian Tamil-language film, directed and produced by B. R. Panthulu. The film stars Sivaj…

1999 single by SlipknotWait and BleedSingle by Slipknotfrom the album Slipknot Released July 1999 February 28, 2000 (commercial)[1] Recorded1998 at Indigo Ranch, Malibu, CaliforniaGenreNu metal[2][3][4]Length2:27LabelRoadrunnerSongwriter(s) Shawn Crahan Chris Fehn Paul Gray Craig Jones Joey Jordison Corey Taylor Mick Thomson Sid Wilson [5] [6]Producer(s) Ross Robinson Slipknot Slipknot singles chronology Wait and Bleed (1999) Spit It Out (2000) Mus…

Gangan humbut adalah makanan yang sering disajikan sebagai menu wajib jika ada hajatan/acara perkawinan/upacara kematian.[1] Gangan humbut merupakan sayur tradisional kalimantan yang memiliki cita rasa yang manis. Makanan ini juga merupakan makanan yang biasa dijual pada bulan Ramadan untuk hidangan berbuka puasa. Cara pembuatan Menggunakan bahan sayur berupa labu dan bahan utamanya batang kelapa muda yang disebut humbut. Diolah dengan ikan gabus atau lebih populer dengan sebutan ikan ha…

这是马来族人名,“莫哈末·雅辛”是父名,不是姓氏,提及此人时应以其自身的名“慕尤丁”为主。 尊敬的丹斯里拿督哈芝慕尤丁·莫哈末雅辛馬來語:Muhyiddin Mohd YassinMahiaddin bin Md Yasin(注册名)国会议员PSM; SPMJ; SHMS; SPSA; SPMP; SUNS; SPDK; DP; PNBS; SMJ; BSI (I); PIS (I)2021年的慕尤丁 第8任马来西亚首相任期2020年3月1日—2021年8月20日君主國家元首蘇丹阿都拉副职依斯迈沙比里前任马哈�…

Irish state agency for the socioeconomic and cultural reinforcement of the Gaeltacht A major contributor to this article appears to have a close connection with its subject. It may require cleanup to comply with Wikipedia's content policies, particularly neutral point of view. Please discuss further on the talk page. (November 2020) (Learn how and when to remove this message) Údarás na GaeltachtaAbbreviationÚnaGFormation1 January 1980FounderIDA IrelandTypeIndustrial developmentIrish languageH…

French title of nobility You can help expand this article with text translated from the corresponding article in French. (October 2022) Click [show] for important translation instructions. Consider adding a topic to this template: there are already 6,196 articles in the main category, and specifying|topic= will aid in categorization. Do not translate text that appears unreliable or low-quality. If possible, verify the text with references provided in the foreign-language article. Y…

30°59′N 6°54′W / 30.983°N 6.900°W / 30.983; -6.900 إقليم ورزازات [[ملف:{{{شعار}}}|200px]] العاصمة ورزازات البلد المغرب الجهة {{{جهة}}} عدد السكان 297.502[1] فردا الإحصاء 2014 المساحة 12.464 كلم² الكثافة 23.87 فرد/كلم² الجهة درعة تافيلالت المقاطعة {{{مقاطعة}}} المطار المدني الأقرب {{{مطار}}} الوالي…